Our APIs
Providing developers and researchers with programmatic access to a vast repository of biodiversity data.

Introduction

Overview
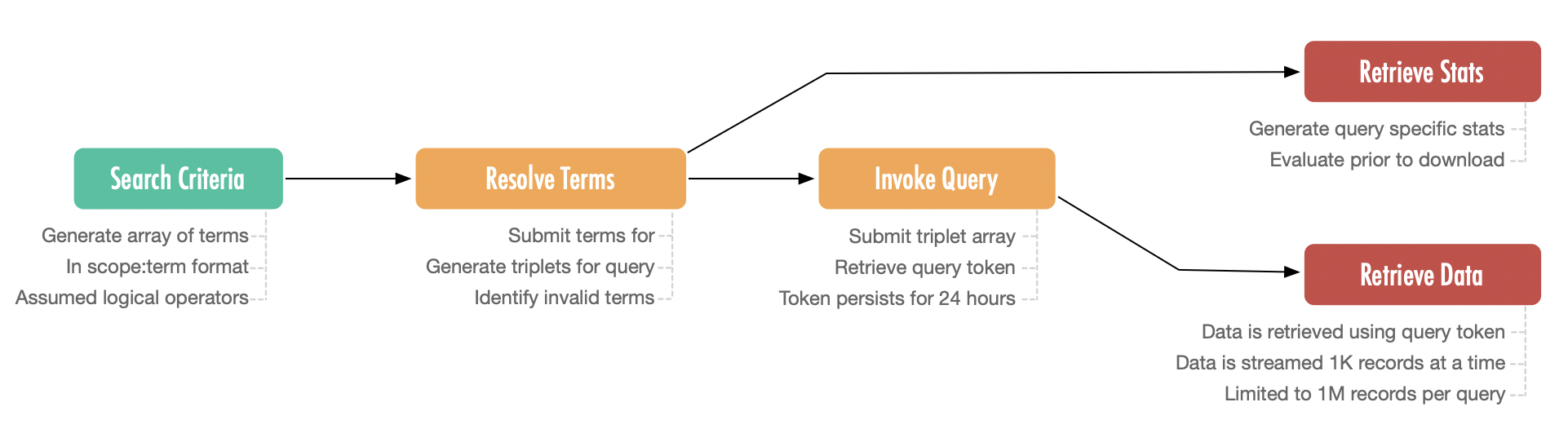
Search Criteria Definition:
Users begin by defining search terms in a standardized scope:term or scope:field:term format. Logical operators are implicitly supported, enabling complex queries. These terms align with controlled vocabularies to ensure consistency and prevent invalid input.
Term Resolution:/api/query/preprocessor
The API validates submitted search terms against controlled vocabularies, flagging invalid entries and generating valid triplets (scope, term, value) for querying. Early validation ensures downstream processes remain error-free.
Query Invocation:/api/query
Validated triplets initiate the query process. A unique query token is generated to reference the query's results. This token remains active for 24 hours, granting users access to associated resources such as statistics or data records.
Retrieve Query Statistics:
(Optional) /api/summary
Users can optionally request summary statistics to evaluate the scope and characteristics of the dataset before retrieving data. This step aids in refining queries and determining the feasibility of large-scale downloads.
Retrieve Data:/api/documents
Data retrieval is conducted in batches of 1,000 records using the query token, with a maximum limit of 1 million records per query. This batching mechanism ensures efficient data streaming, mitigating network timeouts during large downloads and maintaining system scalability.
Preparing Search Queries
The BOLD Portal API enables searching of DNA barcodes by scope and field data that follows the Barcode Core Data Model (BCDM) data standard. Users can use a variety of criteria to prepare a search query in order to dictate the aggregate data and barcodes the API returns to the user, including geographic and taxonomic criteria. Criteria can be combined to allow for different results, where criteria sharing the same scope combine results into a single result, while criteria of different scopes apply an overlap constraint. For example, users can elect to search for 'Aves' barcodes of 'Argentina', which searches for barcodes that overlap on the taxonomic criterion 'Aves' to the geographic criterion 'Argentina'.
The search query is composed of one or more query phrases (split using semicolons) composed of two or three parts: a scope, a field (optional), and a search value. Below is a table of available scopes that can be used to query the API, along with supported optional fields that can be queried for each scope. The query phrase is completed with the value uses would like to query with, and is constructed in the form of <scope>:<value> or <scope>:<field>:<value> if a field is specified.
Using the example above, the query phrase for the taxonomic criterion 'Aves' is tax:Aves or tax:class:Aves, the geopgraphic criterion 'Argentina' is geo:Argentina or geo:country/ocean:Argentina.
Available scopes include:
- tax kingdom,phylum,class,order,family,subfamily,tribe,genus,species,subspecies
Query by taxonomic ranks. - bin uri
Query by BIN. - geo country/ocean,province/state,region
Query by geopolitical divisions. - inst name,seqsite
Query by institution. - recordsetcode code
Query by dataset code. - ids processid,sampleid,insdcacs
Query by ID.
Summary Data
The BOLD Portal API can aggregate barcode metadata into a summary document, as an option to explore the data without committing to performing large queries. Users can specify what metadata will be summarized from the BCDM fields listed below, along with the search query prepared beforehand.
- bin_uri
- coord
- identified_by
- marker_code
- sequence_upload_date
- specimens
- collection_date_start
- country/ocean
- inst
- sequence_run_site
- species
Search queries are run through a preprocessor /api/query/preprocessor to construct a set of formal query triplets. These query triplets are then directed to the /api/summary endpoint to retrieve aggregate barcode metadata.
Using the example above, collecting the summary data for number of specimens in BOLD Portal for the ‘Aves’ of ‘Argentina’ would be as follows:
https://portal.boldsystems.org/api/query/preprocessor?query=tax:Aves;geo:Argentina
https://portal.boldsystems.org/api/summary?query=tax:class:Aves;geo:country/ocean:Argentina&fields=specimensData Retrieval
The BOLD Portal API delivers the DNA barcodes that meets the search query provided by users. The barcodes are returned as documents that follow the BCDM data standard, and users can specify the extent of documents that are returned from the API.
Search queries are run through a preprocessor /api/query/preprocessor to construct a set of formal query triplets. The query triplets, along with the extent of documents to retrieve, are then directed to the /api/query endpoint to retrieve a query_id token. This token is specific to the combination of query triplets and extent specified to the endpoint. Finally, the query_id token is used to download barcodes with /api/documents/<query_id>/download and supports three supported formats: JSON following the BCDM data standard, TSV following the BCDM data standard, or TSV following the Darwin Core data standard.
Only up to 1,000,000 barcodes can be downloaded through the API, so if a selected query exceeds more than 1,000,000 barcodes, a request that includes the query will need to be sent to support@boldsystems.org to download the complete set of data. In the future, an automated system will allow for larger uploads without needing to make a request.
Using the example above, collecting all the barcodes for the 'Aves' of 'Argentina' in the form of JSON documents that follow the BCDM data standard would be as follows:
https://portal.boldsystems.org/api/query/preprocessor?query=tax:Aves;geo:Argentina
https://portal.boldsystems.org/api/query?query=tax:class:Aves;geo:country/ocean:Argentina&extent=full
https://portal.boldsystems.org/api/documents/eAFLT823Ss4vzSspqtTPT05NzLNyLEpPzSvJzEu0LkmssErOSSwutnIsSy22TivNyQEAtOoSIQ==/download?format=jsonFor more detailed documentation on available APIs,
visit the API documentation page.
